About ARGO
ARGO (Aggregate Rank Generator) is a web-based transcriptional regulation resource to aid in the prioritization and interpretation of non-coding disease variants. While many Mendelian diseases are caused by defects in the coding or splicing regions of genes, a precise molecular diagnosis has not been identified for the majority of patients with suspected Mendelian conditions. It is therefore likely that the disease-associated mutations for some of these patients lie in non-coding, regulatory regions of the genome and we need methods for interpreting their functional consequences.
The ENCODE Encyclopedia
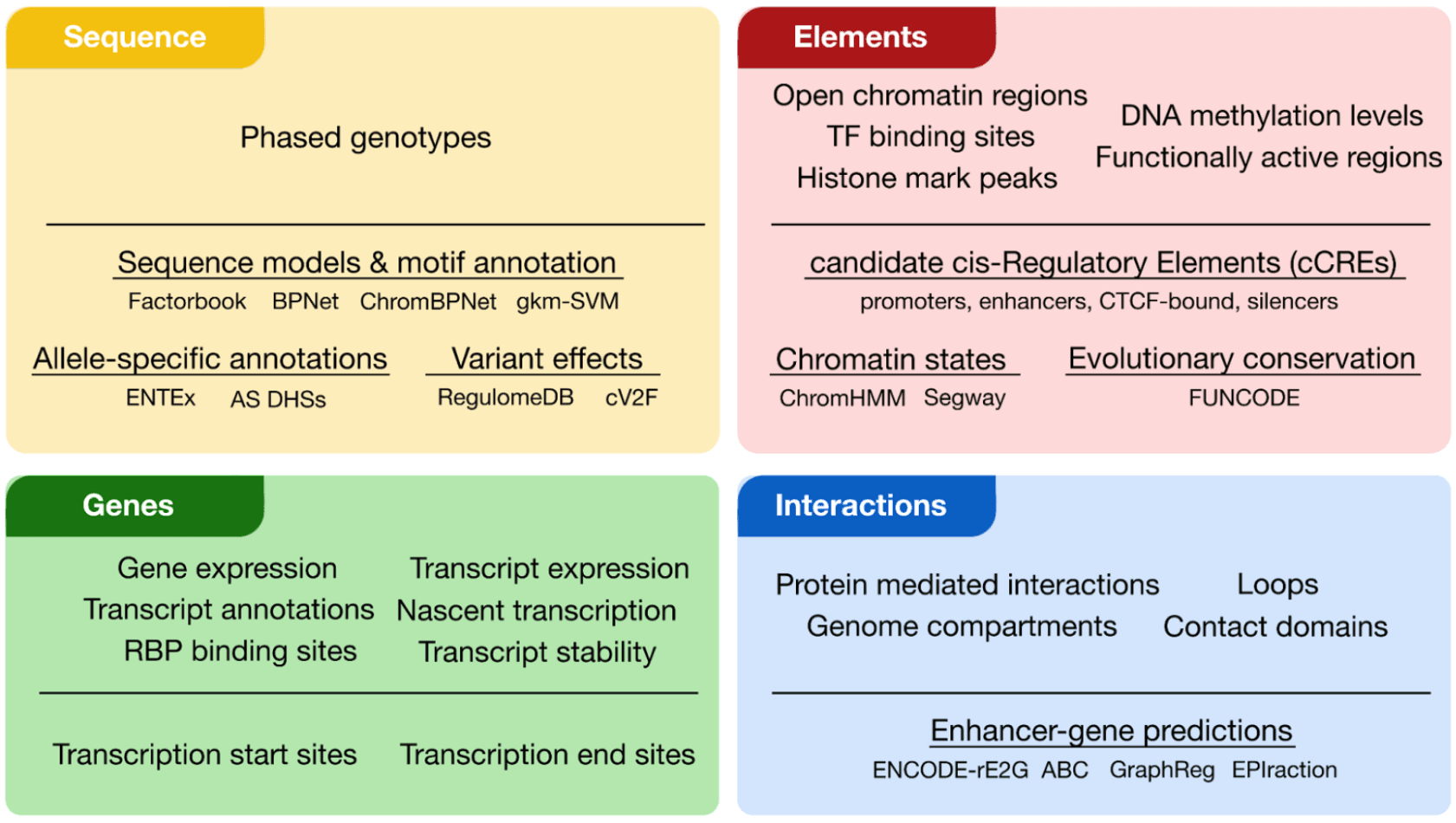
The ENCODE Encyclopedia encompasses a comprehensive set of sequence (yellow), element (red), gene (green) and interaction (blue) annotations (Figure 1). These annotations can either derive directly from primary data (primary level) or derive from the integration of multiple data types using innovative computational methods (integrative level).
Sister Sites
We previously developed a suite of resources for integrating and interpreting regulatory non-coding genomic regions. We led the development of the ENCODE Registry of candidate cis-Regulatory Elements (cCREs), a collection of candidate non-coding regulatory regions across the human and mouse genomes. The Registry contains 2.3 million cCREs in humans and spans thousands of tissues, cell types, and cellular states, comprising promoter, enhancer, silencer, and CTCF-bound elements. To search and visualize these elements, we developed the web portal SCREEN. SCREEN enables users to search for regions, genes, or variants of interest and returns sets of cCREs that can be filtered or visualized based on their predictive functions, activity patterns, and overlap with other genomic annotations (e.g. transcription factor binding sites). In addition to cCRE-centric tools, we have also developed Factorbook2, a web portal that hosts expression data, binding motifs, and binding sites for over 800 transcription factors. These resources provide an integrated platform for researchers to access and interpret data concerning non-coding regulatory regions.
ARGO's purpose is to expand and adapt these tools to develop a comprehensive resource for annotating and prioritizing non-coding variants. We developed a novel variant prioritization scheme which ranks variants based on their overlap with non-coding annotations.

Rank Aggregation
ARGO implements rank aggregation methods, which produce a single consensus ranking by combining multiple different rankings, often deriving from complementary sources. Rank aggregation is advantageous because it can leverage the strengths of individual methods, while mitigating their weaknesses, leading to more robust, reliable, and useful outcomes. Rank aggregation also enables us to integrate diverse data sources with different scales or normalizations as highlighted by our previous work using rank aggregation to predict functionally active enhancers in embryonic mouse tissues.
Annotations
Sequence
ARGO prioritizes candidate variants by aggregating ranks across three categories of annotations: (1) sequence, (2) elements, and (3) genes. For sequence features, variants are ranked based on their predicted impact on transcription factor binding and their evolutionary conservation. Using our Factorbook motif annotations, we calculate the change in motif score between the reference and input variant. To reduce potential false positive motif sites, users are also able to filter motif matches based on support from transcription factor ChIP-seq data. Evolutionary conservation can indicate that a genomic sequence has a crucial biological function and variants in highly conserved regions are more likely to be pathogenic. Because different types of Mendelian conditions may alter regulatory pathways with varying levels of conservation, users can select from conservation scores calculated across vertebrate, mammalian, or primate lineages.
Elements
ARGO also prioritizes variants using element-level annotations by intersecting variants with cCREs. Users have the option to rank elements by their cell type activity and can either broadly group samples by organ or tissue—such as heart, brain, or liver—or select specific cell types such as cardiomyocytes, neurons, or hepatocytes. Users are also able to rank cCREs by the specificity of their activity. This is useful as some conditions may affect multiple systems, suggesting that they disrupt regulatory processes in many different cell types. In contrast, other conditions may be highly specific to a particular cell type, and therefore it is important to prioritize variants that are only active in those cell types
Genes
Finally, ARGO evaluates variants based on properties of their associated genes. To assign variants to target genes, users are able to choose a linking method such as 3D chromatin contacts, computational predictions (ABC4 and ENCODE rE2G5) or linear distance. Then, similar to the cCRE selection, users will then select tissue or cell types of interest for ranking genes by expression as well as denote whether they should include information about the specificity of cell type expression.
